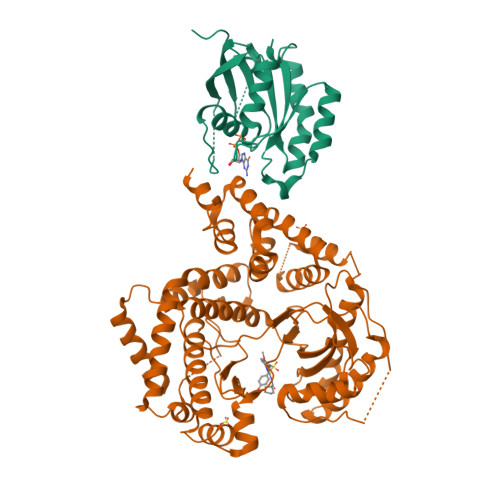
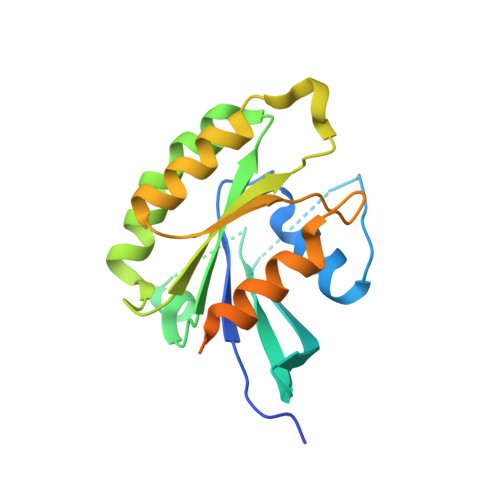
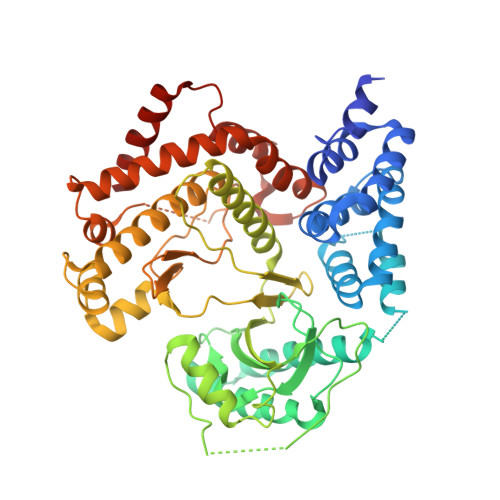
Design and Structural Characterization of Potent and Selective Inhibitors of Phosphatidylinositol 4 Kinase III beta.
Rutaganira, F.U., Fowler, M.L., McPhail, J.A., Gelman, M.A., Nguyen, K., Xiong, A., Dornan, G.L., Tavshanjian, B., Glenn, J.S., Shokat, K.M., Burke, J.E.(2016) J Med Chem 59: 1830-1839
- PubMed: 26885694
- DOI: https://doi.org/10.1021/acs.jmedchem.5b01311
- Primary Citation of Related Structures:
5EUQ - PubMed Abstract:
Type III phosphatidylinositol 4-kinase (PI4KIIIβ) is an essential enzyme in mediating membrane trafficking and is implicated in a variety of pathogenic processes. It is a key host factor mediating replication of RNA viruses. The design of potent and specific inhibitors of this enzyme will be essential to define its cellular roles and may lead to novel antiviral therapeutics. We previously reported the PI4K inhibitor PIK93, and this compound has defined key functions of PI4KIIIβ. However, this compound showed high cross reactivity with class I and III PI3Ks. Using structure-based drug design, we have designed novel potent and selective (>1000-fold over class I and class III PI3Ks) PI4KIIIβ inhibitors. These compounds showed antiviral activity against hepatitis C virus. The co-crystal structure of PI4KIIIβ bound to one of the most potent compounds reveals the molecular basis of specificity. This work will be vital in the design of novel PI4KIIIβ inhibitors, which may play significant roles as antiviral therapeutics.
Organizational Affiliation:
Howard Hughes Medical Institute and Department of Cellular and Molecular Pharmacology, University of California, San Francisco (UCSF) , San Francisco, California 94143, United States.